Two-Group Comparison
Stu Field, SomaLogic Operating Co., Inc.
Source:vignettes/articles/stat-two-group-comparison.Rmd
stat-two-group-comparison.RmdDifferential Expression via t-test
Although targeted statistical analyses are beyond the scope of the
SomaDataIO package, below is an example analysis that
typical users/customers would perform on ‘SomaScan’ data.
It is not intended to be a definitive guide in statistical analysis
and existing packages do exist in the R ecosystem that
perform parts or extensions of these techniques. Many variations of the
workflow below exist, however the framework highlights how one could
perform standard preliminary analyses on ‘SomaScan’ data.
Data Preparation
# the `example_data` .adat object
# download from `SomaLogic-Data` repo or directly via bash command:
# `wget https://raw.githubusercontent.com/SomaLogic/SomaLogic-Data/main/example_data.adat`
# then read in to R with:
# example_data <- read_adat("example_data.adat")
dim(example_data)
#> [1] 192 5318
table(example_data$SampleType)
#>
#> Buffer Calibrator QC Sample
#> 6 10 6 170
# prepare data set for analysis using `preProcessAdat()`
cleanData <- example_data |>
preProcessAdat(
filter.features = TRUE, # remove non-human protein features
filter.controls = TRUE, # remove control samples
filter.rowcheck = TRUE, # retain only passing samples
log.10 = TRUE, # log10 transform
center.scale = FALSE # don't center/scale for univariate analysis
)
#> ✔ 305 non-human protein features were removed.
#> → 214 human proteins did not pass standard QC
#> acceptance criteria and were flagged in `ColCheck`.
#> ✔ 6 buffer samples were removed.
#> ✔ 10 calibrator samples were removed.
#> ✔ 6 QC samples were removed.
#> ✔ 2 samples flagged in `RowCheck` did not
#> pass standard normalization acceptance criteria (0.4 <= x <= 2.5)
#> and were removed.
#> ✔ RFU features were log-10 transformed.
# drop any missing values in Sex
cleanData <- cleanData |>
drop_na(Sex) # rm NAs if present
table(cleanData$Sex)
#>
#> F M
#> 85 83Compare Two Groups (M/F)
Get annotations via getAnalyteInfo():
t_tests <- getAnalyteInfo(cleanData) |>
select(AptName, SeqId, Target = TargetFullName, EntrezGeneSymbol, UniProt)
# Feature data info:
# Subset via dplyr::filter(t_tests, ...) here to
# restrict analysis to only certain analytes
t_tests
#> # A tibble: 4,979 × 5
#> AptName SeqId Target EntrezGeneSymbol UniProt
#> <chr> <chr> <chr> <chr> <chr>
#> 1 seq.10000.28 10000-28 Beta-crystallin B2 CRYBB2 P43320
#> 2 seq.10001.7 10001-7 RAF proto-oncogene… RAF1 P04049
#> 3 seq.10003.15 10003-15 Zinc finger protei… ZNF41 P51814
#> 4 seq.10006.25 10006-25 ETS domain-contain… ELK1 P19419
#> 5 seq.10008.43 10008-43 Guanylyl cyclase-a… GUCA1A P43080
#> 6 seq.10011.65 10011-65 Inositol polyphosp… OCRL Q01968
#> 7 seq.10012.5 10012-5 SAM pointed domain… SPDEF O95238
#> 8 seq.10014.31 10014-31 Zinc finger protei… SNAI2 O43623
#> 9 seq.10015.119 10015-119 Voltage-gated pota… KCNAB2 Q13303
#> 10 seq.10022.207 10022-207 DNA polymerase eta POLH Q9Y253
#> # ℹ 4,969 more rowsCalculate t-tests
Use a “list columns” approach via nested tibble object using
dplyr, purrr, and
stats::t.test()
t_tests <- t_tests |>
mutate(
formula = map(AptName, ~ as.formula(paste(.x, "~ Sex"))), # create formula
t_test = map(formula, ~ stats::t.test(.x, data = cleanData)), # fit t-tests
t_stat = map_dbl(t_test, "statistic"), # pull out t-statistic
p.value = map_dbl(t_test, "p.value"), # pull out p-values
fdr = p.adjust(p.value, method = "BH") # FDR for multiple testing
) |>
arrange(p.value) |> # re-order by `p-value`
mutate(rank = row_number()) # add numeric ranks
# View analysis tibble
t_tests
#> # A tibble: 4,979 × 11
#> AptName SeqId Target EntrezGeneSymbol UniProt formula t_test
#> <chr> <chr> <chr> <chr> <chr> <list> <list>
#> 1 seq.8468.19 8468… Prost… KLK3 P07288 <formula> <htest>
#> 2 seq.6580.29 6580… Pregn… PZP P20742 <formula> <htest>
#> 3 seq.7926.13 7926… Kunit… SPINT3 P49223 <formula> <htest>
#> 4 seq.3032.11 3032… Folli… CGA FSHB P01215… <formula> <htest>
#> 5 seq.16892.23 1689… Ecton… ENPP2 Q13822 <formula> <htest>
#> 6 seq.9282.12 9282… Cyste… CRISP2 P16562 <formula> <htest>
#> 7 seq.5763.67 5763… Beta-… DEFB104A Q8WTQ1 <formula> <htest>
#> 8 seq.2953.31 2953… Lutei… CGA LHB P01215… <formula> <htest>
#> 9 seq.7139.14 7139… SLIT … SLITRK4 Q8IW52 <formula> <htest>
#> 10 seq.4914.10 4914… Human… CGA CGB P01215… <formula> <htest>
#> # ℹ 4,969 more rows
#> # ℹ 4 more variables: t_stat <dbl>, p.value <dbl>, fdr <dbl>,
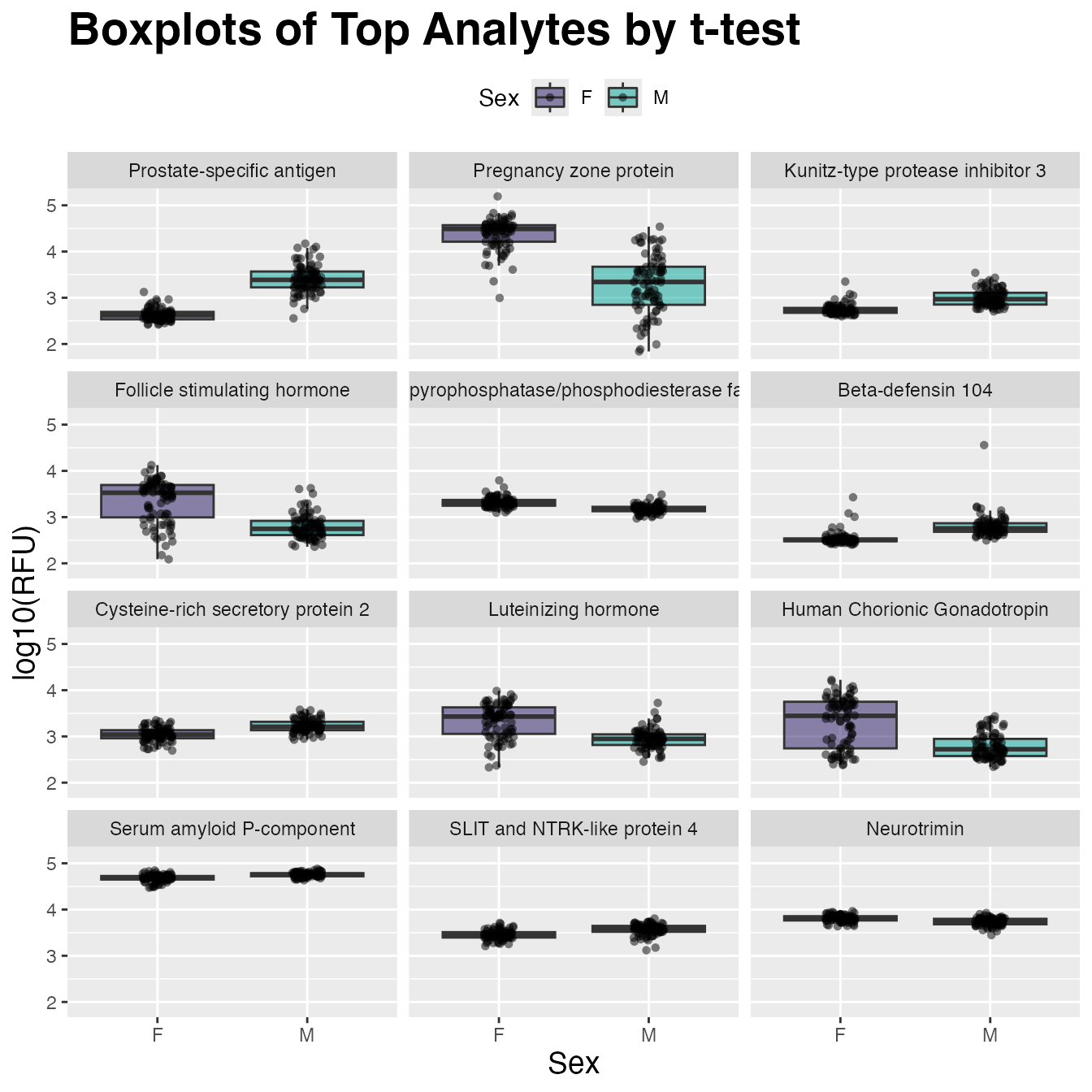
#> # rank <int>Visualize with ggplot2()
Create a plotting tibble in the “long” format for
ggplot2:
target_map <- head(t_tests, 12L) |> # mapping table
select(AptName, Target) # SeqId -> Target
plot_tbl <- example_data |> # plot non-center/scale data
filter(SampleType == "Sample") |> # rm control samples
drop_na(Sex) |> # rm NAs if present
log10() |> # log10-transform for plotting
select(Sex, target_map$AptName) |> # top 12 analytes
pivot_longer(cols = -Sex, names_to = "AptName", values_to = "RFU") |>
left_join(target_map, by = "AptName") |>
# order factor levels by 't_tests' rank to order plots below
mutate(Target = factor(Target, levels = target_map$Target))
plot_tbl
#> # A tibble: 2,040 × 4
#> Sex AptName RFU Target
#> <chr> <chr> <dbl> <fct>
#> 1 F seq.8468.19 2.54 Prostate-specific antigen
#> 2 F seq.6580.29 4.06 Pregnancy zone protein
#> 3 F seq.7926.13 2.66 Kunitz-type protease inhibitor 3
#> 4 F seq.3032.11 3.26 Follicle stimulating hormone
#> 5 F seq.16892.23 3.44 Ectonucleotide pyrophosphatase/phosphodies…
#> 6 F seq.9282.12 2.94 Cysteine-rich secretory protein 2
#> 7 F seq.5763.67 2.52 Beta-defensin 104
#> 8 F seq.2953.31 2.99 Luteinizing hormone
#> 9 F seq.7139.14 3.43 SLIT and NTRK-like protein 4
#> 10 F seq.4914.10 3.93 Human Chorionic Gonadotropin
#> # ℹ 2,030 more rows
plot_tbl |>
ggplot(aes(x = Sex, y = RFU, fill = Sex)) +
geom_boxplot(alpha = 0.5, outlier.shape = NA) +
scale_fill_manual(values = c("#24135F", "#00A499")) +
geom_jitter(shape = 16, width = 0.1, alpha = 0.5) +
facet_wrap(~ Target, ncol = 3) +
ggtitle("Boxplots of Top Analytes by t-test") +
labs(y = "log10(RFU)") +
theme(plot.title = element_text(size = 21, face = "bold"),
axis.title.x = element_text(size = 14),
axis.title.y = element_text(size = 14),
legend.position = "top"
)